We have been working hard to get a release ready for general consumption for the statsmodels code. Well, we're happy to announce that a (very) beta release is ready.
Background
==========
The statsmodels code was started by Jonathan Taylor and was formerly included as part of scipy. It was taken up to be tested, corrected, and extended as part of the Google Summer of Code 2009.
What it is
==========
We are now releasing the efforts of the last few months under the scikits namespace as scikits.statsmodels. Statsmodels is a pure python package that requires numpy and scipy. It offers a convenient interface for fitting parameterized statistical models with growing support for displaying univariate and multivariate summary statistics, regression summaries, and (postestimation) statistical tests.
Main Feautures
==============
* regression: Generalized least squares (including weighted least squares and least squares with autoregressive errors), ordinary least squares.
* glm: Generalized linear models with support for all of the one-parameter exponential family distributions.
* rlm: Robust linear models with support for several M-estimators.
* datasets: Datasets to be distributed and used for examples and in testing.
There is also a sandbox which contains code for generalized additive models (untested), mixed effects models, cox proportional hazards model (both are untested and still dependent on the nipy formula framework), generating descriptive statistics, and printing table output to ascii, latex, and html. None of this code is considered "production ready".
Where to get it
===============
Development branches will be on LaunchPad. This is where to go to get the most up to date code in the trunk branch. Experimental code will also be hosted here in different branches.
https://code.launchpad.net/statsmodels
Source download of stable tags will be on SourceForge.
https://sourceforge.net/projects/statsmodels/
or
PyPi: http://pypi.python.org/pypi/scikits.statsmodels/
License
=======
Simplified BSD
Documentation
=============
The official documentation is hosted on SourceForge.
http://statsmodels.sourceforge.net/
The sphinx docs are currently undergoing a lot of work. They are not yet comprehensive, but should get you started.
This blog will continue to be updated as we make progress on the code.
Discussion and Development
==========================
All chatter will take place on the or scipy-user mailing list. We are very interested in receiving feedback about usability, suggestions for improvements, and bug reports via the mailing list or the bug tracker at https://bugs.launchpad.net/statsmodels.
Monday, August 31, 2009
Thursday, August 27, 2009
GSoC Is Over
Whoa, where did the last month go? The Google Summer of Code 2009 officially ended this Monday. Though I haven't taken a breath to update the blog, we (Josef and I) have been hard at work on the models code.
We have working and tested versions of Generalized Least Squares, Weighted Least Squares, Ordinary Least Squares, Robust Linear Models with several M-estimators, and Generalized Linear Models with support for all (almost all?) one parameter exponential family distributions. We have also provided some more convenience functions, created a standalone python package for the models code, and obtained permissions to distribute a few more datasets. Due to a lack of time, there is only experimental (read untested) support for autoregressive models, mixed effects models, generalized additive models, and convenience functions for returning strings (possibly html and latex output as well) with regression results and descriptive statistics. I will continue to work on these as I find time.
I will soon post a note on the progress that was made in the robust linear models code. Also, look out for a (semi-) official release of the code in the next few days. We have decided to name the project statsmodels and distribute it as a scikit. We need to finalize the documentation (should be ready to go in the next day or so...I am back taking courses) and clean up some of the usage examples, so people can jump right in and use the code, give feedback, and hopefully contribute extensions and new models.
As for the future of statsmodels, we are discussing over the next few weeks the immediate extensions that we know would like to make. It's looking like I will be wearing my microeconometrician hat this semester in my own coursework. More specifically, I will probably be working with cross-sectional and panel data models for household survey data in my own research and finding some time for time series models as part of my teaching assistantship. Josef has also mentioned wanting to work more with time series models.
If anyone (especially those from other disciplines) would like to contribute or see some extensions (my apologies to those who have made requests that I haven't yet been able to accomodate) feel free to post to the scipy-dev mailing list. I'm more than happy to discuss/debate with users and potential developers the design decisions that have been made, as I think the code is still in an unsettled enough state to merit some discussion.
We have working and tested versions of Generalized Least Squares, Weighted Least Squares, Ordinary Least Squares, Robust Linear Models with several M-estimators, and Generalized Linear Models with support for all (almost all?) one parameter exponential family distributions. We have also provided some more convenience functions, created a standalone python package for the models code, and obtained permissions to distribute a few more datasets. Due to a lack of time, there is only experimental (read untested) support for autoregressive models, mixed effects models, generalized additive models, and convenience functions for returning strings (possibly html and latex output as well) with regression results and descriptive statistics. I will continue to work on these as I find time.
I will soon post a note on the progress that was made in the robust linear models code. Also, look out for a (semi-) official release of the code in the next few days. We have decided to name the project statsmodels and distribute it as a scikit. We need to finalize the documentation (should be ready to go in the next day or so...I am back taking courses) and clean up some of the usage examples, so people can jump right in and use the code, give feedback, and hopefully contribute extensions and new models.
As for the future of statsmodels, we are discussing over the next few weeks the immediate extensions that we know would like to make. It's looking like I will be wearing my microeconometrician hat this semester in my own coursework. More specifically, I will probably be working with cross-sectional and panel data models for household survey data in my own research and finding some time for time series models as part of my teaching assistantship. Josef has also mentioned wanting to work more with time series models.
If anyone (especially those from other disciplines) would like to contribute or see some extensions (my apologies to those who have made requests that I haven't yet been able to accomodate) feel free to post to the scipy-dev mailing list. I'm more than happy to discuss/debate with users and potential developers the design decisions that have been made, as I think the code is still in an unsettled enough state to merit some discussion.
Saturday, July 25, 2009
Iterated Reweighted Least Squares
I have spent the last two weeks putting the "finishing" touches on the generalized linear models and starting to go over the robust linear models (RLM). The test suite for GLM is not complete yet, but all of the exponential families are covered with at least their default link functions tested and are looking good. So in an effort to make a first pass over all of the existing code, I moved on to RLM. After the time spent with GLM theory, the RLMs theory and code was much more manageable.
Before discussing the RLMs, their implementation, and the extensions I have made. I will describe the iterated reweighted least squares (IRLS) algorithm for the GLMs to demonstrate the theory and the solution method in the models code. A very similar iteration is done for the RLMs as well.
The main idea of GLM, as noted, is to relate a response variable to a linear model via a link function, which allows us to use least squares regression. Let us take as an example, the binomial family (which is written to handle Bernoulli and binomial data). In this case, the response variable is Bernoulli, 1 indicates a "success" and 0 a "failure".
The default link for the binomial family is the logit link. So we have
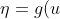=\ln\frac{\mu}{1-\mu})
η is our linear predictor and μ is our actual mean response. The first thing that we need for the algorithm is to compute a first guess on μ (IRLS as opposed to Newton-Raphson makes a guess on the mean response rather than the parameter vector β). The algorithm is fairly robust to this first guess; however, a common choice is
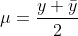
For the binomial family, we specifically use
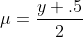
where y is our given response variable. We then use this first guess to initialize our linear predictor η via the link function given above. With these estimates we are able to start the iteration. Our convergence criteria is based on the deviance function, which is simply twice the log-likelihood ratio of our current guess on the fitted model versus the saturated log-likelihood.
![D=2\phi\left[ \mathcal{L}\left(y;y \right ) - \mathcal{L}\left(y;\mu \right )\right ]](http://latex.codecogs.com/gif.latex?D=2\phi\left[ \mathcal{L}\left(y;y \right ) - \mathcal{L}\left(y;\mu \right )\right ])
Where Φ is a dispersion (scale) parameter. Note that the saturated log-likelihood is simply the likelihood of the perfectly fitted model where y = μ. For the binomial family the deviance function is
 & \text{, if }y_{i}=0\\ -2\ln\mu_{i} & \text{, if }y_{i}=1\end{cases})
The iteration continues while the deviance function evaluated at the updated μ differs from the previous by the given convergence tolerance (default is 1e-08) and the number of iterations is less than the given maximum (default is 100).
The actual iterations, as the name of the algorithm suggests, run a weighted least squares fit of the actual regressors on the adjusted linear predictor (our transformed guess on the response variable). The adjustment is given by
 \\ \text{Recalling that }\eta=g(\mu))
which moves us closer to the root of the estimating equation (see the previously mentioned Gill or Hardin and Hilbe for the details of root-finding using the Newton-Raphson method. IRLS is simply Newton-Raphson using some simplifications.). The only remaining ingredient is the choice of weights for the weighted least squares. Similar to other methods such as general least squares (GLS) that correct for heteroskedasticity in the error terms, a diagonal matrix containing estimates of the variance of the error terms is used. However, in our case, the exact form of this variance is unknown and difficult to estimate, but the error each estimate is assumed to vary with the mean response variable. Thus, we improve the weights at each step given our best guess for the mean response variable μ and the known variance function of each family. For the binomial family, as is well known, the variance function is
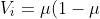)
Thus we can obtain an estimate of the parameters using weighted least squares. Using these estimates we update η
)
Correcting by an offset if one is specified (the code currently does not support the use of offsets, but this would be a simple extension). Using this linear predictor (remember it was originally given by the linear transformation η = g(μ)) we update our guess on the mean response variable μ and use this to update our estimate of the deviance function. This is continued until convergence is reached.
Once the fit is obtained, the covariance matrix is obtained as always though in the case of GLM it is weighted by an estimate of the scale chosen when the fit method is called. The default is Pearson's Chi-Squared. The standard errors of the estimate are obtained from the diagonal elements of this matrix.
And that's basically it. Much of my time with the GLM code was spent getting my head around the theory and then this algorithm. Then I had to obtain data (Jeff Gill was kind enough to give permission for SciPy to use and distribute the datasets from his nice monograph) and write tests to ensure that all of the intermediate and final results were correct for each family. This was no small feat considering there are 6 families after I added the negative binomial family and around 25 possible combinations of families and link functions. Figuring out the correct use of the estimated scale (or dispersion) parameters for each family was particularly challenging. As I mentioned, in the interest of time, I haven't written tests for the noncanonical links for each and every family, but the initial results look good and these tests will come.
GLM provided a good base for understanding the remaining code and allowed me to more or less plow through the robust linear models estimator. I had some mathematical difficulties in extending the models to include the correct covariance matrices, since there is no strong theoretical consensus on what they should actually be! More on RLM in my next post, but before then I'll just give a quick look at how the GLM estimator is used.
First, the algorithm described above has been made flexible enough to estimate truly binomial data. That is, we can have a vector with each row containing (# of successes, # of failures) as is the case in the star98 data from Dr. Gill, described and included in the models.datasets. It will be useful to have a look at the syntax for this type of data as it's slightly different than the other families.
In [1]: import models
In [2]: import numpy as np
In [3]: from models.datasets.star98.data import load
In [4]: data = load()
In [5]: data.exog = models.functions.add_constant(data.exog)
In [6]: trials = data.endog[:,:2].sum(axis=1)
In [7]: data.endog[0] # (successes, failures)
Out[7]: array([ 452., 355.])
In [8]: trials[0] # total trials
Out[8]: 807.0
In [9]: from models.glm import GLMtwo as GLM # the name will change
In [10]: bin_model = GLM(data.endog, data.exog, family=models.family.Binomial())
In [11]: bin_results = bin_model.fit(data_weights = trials)
In [12]: bin_results.params
Out[12]:
array([ -1.68150366e-02, 9.92547661e-03, -1.87242148e-02,
-1.42385609e-02, 2.54487173e-01, 2.40693664e-01,
8.04086739e-02, -1.95216050e+00, -3.34086475e-01,
-1.69022168e-01, 4.91670212e-03, -3.57996435e-03,
-1.40765648e-02, -4.00499176e-03, -3.90639579e-03,
9.17143006e-02, 4.89898381e-02, 8.04073890e-03,
2.22009503e-04, -2.24924861e-03, 2.95887793e+00])
The main difference between the above and the rest of the families is that you must specify the total number of "trials" (which as you can see is just the sum of success and failures for each observation) as the data_weights argument to fit. This was done so that the current implementation of the Bernouilli family could be extended without rewriting its other derived functions. The code could easily (and might be) extended to calculate these trials so that this argument doesn't need to be specified, but it's sometimes better to be explicit.
Before discussing the RLMs, their implementation, and the extensions I have made. I will describe the iterated reweighted least squares (IRLS) algorithm for the GLMs to demonstrate the theory and the solution method in the models code. A very similar iteration is done for the RLMs as well.
The main idea of GLM, as noted, is to relate a response variable to a linear model via a link function, which allows us to use least squares regression. Let us take as an example, the binomial family (which is written to handle Bernoulli and binomial data). In this case, the response variable is Bernoulli, 1 indicates a "success" and 0 a "failure".
The default link for the binomial family is the logit link. So we have
η is our linear predictor and μ is our actual mean response. The first thing that we need for the algorithm is to compute a first guess on μ (IRLS as opposed to Newton-Raphson makes a guess on the mean response rather than the parameter vector β). The algorithm is fairly robust to this first guess; however, a common choice is
For the binomial family, we specifically use
where y is our given response variable. We then use this first guess to initialize our linear predictor η via the link function given above. With these estimates we are able to start the iteration. Our convergence criteria is based on the deviance function, which is simply twice the log-likelihood ratio of our current guess on the fitted model versus the saturated log-likelihood.
Where Φ is a dispersion (scale) parameter. Note that the saturated log-likelihood is simply the likelihood of the perfectly fitted model where y = μ. For the binomial family the deviance function is
The iteration continues while the deviance function evaluated at the updated μ differs from the previous by the given convergence tolerance (default is 1e-08) and the number of iterations is less than the given maximum (default is 100).
The actual iterations, as the name of the algorithm suggests, run a weighted least squares fit of the actual regressors on the adjusted linear predictor (our transformed guess on the response variable). The adjustment is given by
which moves us closer to the root of the estimating equation (see the previously mentioned Gill or Hardin and Hilbe for the details of root-finding using the Newton-Raphson method. IRLS is simply Newton-Raphson using some simplifications.). The only remaining ingredient is the choice of weights for the weighted least squares. Similar to other methods such as general least squares (GLS) that correct for heteroskedasticity in the error terms, a diagonal matrix containing estimates of the variance of the error terms is used. However, in our case, the exact form of this variance is unknown and difficult to estimate, but the error each estimate is assumed to vary with the mean response variable. Thus, we improve the weights at each step given our best guess for the mean response variable μ and the known variance function of each family. For the binomial family, as is well known, the variance function is
Thus we can obtain an estimate of the parameters using weighted least squares. Using these estimates we update η
Correcting by an offset if one is specified (the code currently does not support the use of offsets, but this would be a simple extension). Using this linear predictor (remember it was originally given by the linear transformation η = g(μ)) we update our guess on the mean response variable μ and use this to update our estimate of the deviance function. This is continued until convergence is reached.
Once the fit is obtained, the covariance matrix is obtained as always though in the case of GLM it is weighted by an estimate of the scale chosen when the fit method is called. The default is Pearson's Chi-Squared. The standard errors of the estimate are obtained from the diagonal elements of this matrix.
And that's basically it. Much of my time with the GLM code was spent getting my head around the theory and then this algorithm. Then I had to obtain data (Jeff Gill was kind enough to give permission for SciPy to use and distribute the datasets from his nice monograph) and write tests to ensure that all of the intermediate and final results were correct for each family. This was no small feat considering there are 6 families after I added the negative binomial family and around 25 possible combinations of families and link functions. Figuring out the correct use of the estimated scale (or dispersion) parameters for each family was particularly challenging. As I mentioned, in the interest of time, I haven't written tests for the noncanonical links for each and every family, but the initial results look good and these tests will come.
GLM provided a good base for understanding the remaining code and allowed me to more or less plow through the robust linear models estimator. I had some mathematical difficulties in extending the models to include the correct covariance matrices, since there is no strong theoretical consensus on what they should actually be! More on RLM in my next post, but before then I'll just give a quick look at how the GLM estimator is used.
First, the algorithm described above has been made flexible enough to estimate truly binomial data. That is, we can have a vector with each row containing (# of successes, # of failures) as is the case in the star98 data from Dr. Gill, described and included in the models.datasets. It will be useful to have a look at the syntax for this type of data as it's slightly different than the other families.
In [1]: import models
In [2]: import numpy as np
In [3]: from models.datasets.star98.data import load
In [4]: data = load()
In [5]: data.exog = models.functions.add_constant(data.exog)
In [6]: trials = data.endog[:,:2].sum(axis=1)
In [7]: data.endog[0] # (successes, failures)
Out[7]: array([ 452., 355.])
In [8]: trials[0] # total trials
Out[8]: 807.0
In [9]: from models.glm import GLMtwo as GLM # the name will change
In [10]: bin_model = GLM(data.endog, data.exog, family=models.family.Binomial())
In [11]: bin_results = bin_model.fit(data_weights = trials)
In [12]: bin_results.params
Out[12]:
array([ -1.68150366e-02, 9.92547661e-03, -1.87242148e-02,
-1.42385609e-02, 2.54487173e-01, 2.40693664e-01,
8.04086739e-02, -1.95216050e+00, -3.34086475e-01,
-1.69022168e-01, 4.91670212e-03, -3.57996435e-03,
-1.40765648e-02, -4.00499176e-03, -3.90639579e-03,
9.17143006e-02, 4.89898381e-02, 8.04073890e-03,
2.22009503e-04, -2.24924861e-03, 2.95887793e+00])
The main difference between the above and the rest of the families is that you must specify the total number of "trials" (which as you can see is just the sum of success and failures for each observation) as the data_weights argument to fit. This was done so that the current implementation of the Bernouilli family could be extended without rewriting its other derived functions. The code could easily (and might be) extended to calculate these trials so that this argument doesn't need to be specified, but it's sometimes better to be explicit.
Saturday, July 11, 2009
GLM Residuals and The Beauty of Stats with Python + SciPy
I just finished including the Anscombe residuals for the families in the generalized linear models. The Anscombe residuals for the Binomial family were particularly tricky. It took me a while to work through the math and then figure out the SciPy syntax for what I need (some docs clarification coming...), but if I had had to implement these functions myself (presumably not in Python), it would have taken me more than a week! I thought it provided a good opportunity introduce the residuals and to show off how easy things are in Python with NumPy/SciPy.
Note that there is not really a unified terminology for residual analysis in GLMs. I will try to use the most common names for these residuals in introducing the basics and point out any deviations from the norm both here and in the SciPy documentation.
In general, residuals should be defined such that their distribution is approximately normal. This is achieved through the use of linear equations, transformed linear equations, and deviance contributions. Below you will find the five most common types of residuals that rely mainly on transformations and one that relies on deviance contributions, though there are as many as 9+ different types of residuals in the literature.
These are simply the observed response values minus the predicted values.

In a classic linear model these are just the expected residuals

However, for GLM, these become

where is the link function that makes our model's linear predictor comparable to response vector.
is the link function that makes our model's linear predictor comparable to response vector.
It is, however, common in GLMs to produce residuals that deviate greatly from the classical assumptions needed in residual analysis, so in addition we have these other residuals which attempt to mitigate deviations from the needed assumptions.
The Pearson residuals are a version of the response residuals, scaled by the standard deviation of the prediction. The name comes from the fact that the sum of the Pearson residuals for a Poisson GLM is equal to Pearson's statistic, a goodness of fit measure.
statistic, a goodness of fit measure.
![r_{i}^{P}=\frac{(y_{i}-\hat{\mu_{i}})}{\sqrt{\text{VAR}[\hat{\mu}_{i}]}}](http://codecogs.izyba.com/gif.latex?r_%7Bi%7D%5E%7BP%7D=%5Cfrac%7B%28y_%7Bi%7D-%5Chat%7B%5Cmu_%7Bi%7D%7D%29%7D%7B%5Csqrt%7B%5Ctext%7BVAR%7D%5B%5Chat%7B%5Cmu%7D_%7Bi%7D%5D%7D%7D)
The scaling allows plotting of these residuals versus an individual predictor or the outcome to identify outliers. The problem with these residuals though is that asymptotically they are normally distributed, but in practice they can be quite skewed leading to a misinterpretation of the actual dispersion.
These are the difference between the working response and the linear predictor at convergence (ie., the last step in the iterative process).
\left.\left(\frac{\partial\eta}{\partial\mu}\right)_{i}\right|_{\mu=\hat{\mu}})
Anscombe (1960, 1961) describes a general transformation in place of
in place of  so that they are as close to a normal distribution as possible. (Note: "There is a maddeningly great diversity of the forms that the Anscombe residuals take in the literature." I have included the simplest as described below. (Gill 54, emphasis added)) The function
so that they are as close to a normal distribution as possible. (Note: "There is a maddeningly great diversity of the forms that the Anscombe residuals take in the literature." I have included the simplest as described below. (Gill 54, emphasis added)) The function  is given by
is given by
![A(\cdot)=\int\text{VAR}[\mu]^{-\frac{1}{3}}d\mu](http://codecogs.izyba.com/gif.latex?A%28%5Ccdot%29=%5Cint%5Ctext%7BVAR%7D%5B%5Cmu%5D%5E%7B-%5Cfrac%7B1%7D%7B3%7D%7Dd%5Cmu)
This is done for both the response and the predictions. This difference is then scaled by dividing by

so that the Anscombe Residuals are
-A\left(\hat{\mu}_{i} \right )}{A^{\prime}\left(\hat{\mu}_{i}\right)\sqrt{\text{VAR}\left(\hat{\mu}_{i} \right )} \right )})
The Poisson distribution has constant variance so that it's Anscombe Residuals are simply
so that it's Anscombe Residuals are simply

Easy right? Sure was until I ran into the binomial distribution. The Anscombe residuals are built up in a different way for the binomial family. Indeed, the McCullagh and Nelder text does not even provide the general formula for the binomial Anscombe residuals and refers the reader to Anscombe (1953) and Cox & Snell (1968). The problem is that following this transformation for the binomial distribution leads to a rather nasty solution involving the hypergeometric 2F1 function or equivalently the incomplete beta function multiplied by the beta function as shown by Cox and Snell (1968).
Gill writes "A partial reason that Anscombe residuals are less popular than other forms is the difficulty in obtaining these tabular values." The tabular values to which he is referring are found in Cox and Snell (1968) p. 260. It is a table of the incomplete beta function that was tabulated numerically for an easy reference. How difficult would it be to get this table with NumPy and SciPy?
import numpy as np
from scipy import special
betainc = lambda x: special.betainc(2/3.,2/3.,x)
table = np.arange(0,.5,.001).reshape(50,10)
results = []
for i in table:
results.append(betainc(i))
results = np.asarray(results).reshape(50,10)
That's it!
Now the Anscombe residuals for the binomial distribution are

Where n is the number of trials for each observation (ie., 1, , or
, or  )
)
To implement this in the GLM binomial family class, I defined an intermediate function cox_snell similar to the above. It now looks like
from scipy import special
import numpy as np
def resid_anscombe(self, Y, mu):
cox_snell = lambda x: special.betainc(2/3.,2/3.,x)*special.beta(2/3.,2/3.)
return np.sqrt(self.n)*(cox_snell(Y)-cox_snell(mu))/(mu**(1/6.)*(1-mu)**(1/6.))
We multiply the above formula by np.sqrt(self.n) the square root of the number of trials in order to correct for possible use of proportional outcomes Y and mu.
Also, see the ticket here for a note about the incomplete beta function.
One other important type of residual in GLMs is the deviance residual. The deviance residual is the most general and also the most useful of the GLM residuals. The IRLS algorithm (as will be shown in a future post) depends on the convergence of the deviance function. The deviance residual then is just the increment to the overall deviance of each observation.

where are defined for each family.
are defined for each family.
Note that most of these residuals also come in variations such as modified, standardized, studentized, and adjusted.
Anscombe, FJ (1953) "Contribution to the discussion of H. Hotelling's paper."
Journal of the Royal Statistical Society, B, 15, 229-30.
Anscombe, FJ (1960) "Rejection of outliers." Technometrics, 2,
123-47.
Anscombe, FJ (1961) "Examination of residuals." Proceedings of
the Fourth Berkeley Symposium on Mathematical Statistics and
Probability. Berkeley: University of California Press.
Cox, DR and Snell, EJ (1968). "A generalized definition of
residuals." Journal of the Royal Statistical Society B, 30, 248-65.
Note that there is not really a unified terminology for residual analysis in GLMs. I will try to use the most common names for these residuals in introducing the basics and point out any deviations from the norm both here and in the SciPy documentation.
In general, residuals should be defined such that their distribution is approximately normal. This is achieved through the use of linear equations, transformed linear equations, and deviance contributions. Below you will find the five most common types of residuals that rely mainly on transformations and one that relies on deviance contributions, though there are as many as 9+ different types of residuals in the literature.
Response Residuals
These are simply the observed response values minus the predicted values.
In a classic linear model these are just the expected residuals
However, for GLM, these become
where
It is, however, common in GLMs to produce residuals that deviate greatly from the classical assumptions needed in residual analysis, so in addition we have these other residuals which attempt to mitigate deviations from the needed assumptions.
Pearson Residuals
The Pearson residuals are a version of the response residuals, scaled by the standard deviation of the prediction. The name comes from the fact that the sum of the Pearson residuals for a Poisson GLM is equal to Pearson's
The scaling allows plotting of these residuals versus an individual predictor or the outcome to identify outliers. The problem with these residuals though is that asymptotically they are normally distributed, but in practice they can be quite skewed leading to a misinterpretation of the actual dispersion.
Working Residuals
These are the difference between the working response and the linear predictor at convergence (ie., the last step in the iterative process).
Anscombe Residuals
Anscombe (1960, 1961) describes a general transformation
This is done for both the response and the predictions. This difference is then scaled by dividing by
so that the Anscombe Residuals are
The Poisson distribution has constant variance
Easy right? Sure was until I ran into the binomial distribution. The Anscombe residuals are built up in a different way for the binomial family. Indeed, the McCullagh and Nelder text does not even provide the general formula for the binomial Anscombe residuals and refers the reader to Anscombe (1953) and Cox & Snell (1968). The problem is that following this transformation for the binomial distribution leads to a rather nasty solution involving the hypergeometric 2F1 function or equivalently the incomplete beta function multiplied by the beta function as shown by Cox and Snell (1968).
Gill writes "A partial reason that Anscombe residuals are less popular than other forms is the difficulty in obtaining these tabular values." The tabular values to which he is referring are found in Cox and Snell (1968) p. 260. It is a table of the incomplete beta function that was tabulated numerically for an easy reference. How difficult would it be to get this table with NumPy and SciPy?
import numpy as np
from scipy import special
betainc = lambda x: special.betainc(2/3.,2/3.,x)
table = np.arange(0,.5,.001).reshape(50,10)
results = []
for i in table:
results.append(betainc(i))
results = np.asarray(results).reshape(50,10)
That's it!
Now the Anscombe residuals for the binomial distribution are
Where n is the number of trials for each observation (ie., 1,
To implement this in the GLM binomial family class, I defined an intermediate function cox_snell similar to the above. It now looks like
from scipy import special
import numpy as np
def resid_anscombe(self, Y, mu):
cox_snell = lambda x: special.betainc(2/3.,2/3.,x)*special.beta(2/3.,2/3.)
return np.sqrt(self.n)*(cox_snell(Y)-cox_snell(mu))/(mu**(1/6.)*(1-mu)**(1/6.))
We multiply the above formula by np.sqrt(self.n) the square root of the number of trials in order to correct for possible use of proportional outcomes Y and mu.
Also, see the ticket here for a note about the incomplete beta function.
Deviance Residuals
One other important type of residual in GLMs is the deviance residual. The deviance residual is the most general and also the most useful of the GLM residuals. The IRLS algorithm (as will be shown in a future post) depends on the convergence of the deviance function. The deviance residual then is just the increment to the overall deviance of each observation.
where
Note that most of these residuals also come in variations such as modified, standardized, studentized, and adjusted.
Selected References
Anscombe, FJ (1953) "Contribution to the discussion of H. Hotelling's paper."
Journal of the Royal Statistical Society, B, 15, 229-30.
Anscombe, FJ (1960) "Rejection of outliers." Technometrics, 2,
123-47.
Anscombe, FJ (1961) "Examination of residuals." Proceedings of
the Fourth Berkeley Symposium on Mathematical Statistics and
Probability. Berkeley: University of California Press.
Cox, DR and Snell, EJ (1968). "A generalized definition of
residuals." Journal of the Royal Statistical Society B, 30, 248-65.
Test Post
I am testing out my new ASCIIMathML script, so I can type LaTeX in blogger.
Note that to correctly view this page requires Internet Explorer 6 + MathPlayer or Firefox/Mozilla/Netscape (with MathML fonts). So the equations won't work in a feed reader.
$A(\cdot)=\int\,$VAR$\left[\mu\right]^{-\frac{1}{3}}\, d\mu$
Well it appears to work pretty well, except the \text{} tag doesn't work. Hmm, though it should.
This works.
amath
` text(TEXT) `
endamath
This doesn't work.
$\text{TEXT}$
You can get the ASCIIMathML script here and follow these directions to add a javascript widget.
Another possible solution for typing equations in Blogger relies on typing LaTeX equations into a remote site. This didn't really appeal to me, as I don't foresee needing any overly complicated mathematical formulae, though it seems like a perfectly workable solution. You can read about it here
[Edit: I might switch to this approach, since the equations don't display if you don't have the above mentioned requirements or are in a feed reader.]
Hat tip to Please Make a Note for the pointers.
Note that to correctly view this page requires Internet Explorer 6 + MathPlayer or Firefox/Mozilla/Netscape (with MathML fonts). So the equations won't work in a feed reader.
$A(\cdot)=\int\,$VAR$\left[\mu\right]^{-\frac{1}{3}}\, d\mu$
Well it appears to work pretty well, except the \text{} tag doesn't work. Hmm, though it should.
This works.
amath
` text(TEXT) `
endamath
This doesn't work.
$\text{TEXT}$
You can get the ASCIIMathML script here and follow these directions to add a javascript widget.
Another possible solution for typing equations in Blogger relies on typing LaTeX equations into a remote site. This didn't really appeal to me, as I don't foresee needing any overly complicated mathematical formulae, though it seems like a perfectly workable solution. You can read about it here
[Edit: I might switch to this approach, since the equations don't display if you don't have the above mentioned requirements or are in a feed reader.]
Hat tip to Please Make a Note for the pointers.
Thursday, July 2, 2009
Generalized Linear Models
As I have mentioned, I have spent the last few weeks both in stats books, finding my way around R, and cleaning up and refactoring the code for the generalized linear models in the NiPy models code. I have recently hit a wall in this code, so I am trying to clear out some unposted blog drafts. I intended for this post to introduce the generalized linear models approach to estimation; however the full post will have to wait. For now, I will give an introduction to the theory and then explain where I am with the code.
Generalized linear models was a topic that was completely foreign to me a few weeks ago, but after a little (okay a lot of) reading the approach seems almost natural. I have found the following references useful:
The basic point of the generalized linear model is to extend the approach taken in classical linear regression to models that have more complex outcomes but ultimately share the linearity property. In this respect, GLM subsumes classical linear regression, probit and logit analysis, loglinear and multinomial response models, and some models that deal with survival data to name a few.
In my experience, I have found that econometrics is taught in a compartmentalized manner. This makes sense to a certain extent, as different estimators are tailored to particular problems and data. GLM on the other hand allows the use of a common a technique for obtaining parameter estimates so that it can be studied as a single technique rather than as a collection of distinct approaches.
If interested in my ramblings, you can find a draft of my notes as an introduction to GLM here, as Blogger does not support LaTeX... Please note that this is a preliminary and incomplete draft (corrections and clarifications are very welcome). One thing it could definitely use is some clarification by example. However, as I noted, I have run into a bit of a wall trying to extend the binomial family to accept a vector of proportional data, and this is my intended example to walk through the theory and algorithm, so... a subsequent post will have to lay this out once I've got it sorted myself.
Generally speaking, there are two basic algorithms for GLM estimation: one is a maximum likelihood optimization based on Newton's method the other is commonly refered to as iteratively (re)weighted least squares (IRLS or IWLS). Our implementation now only covers IRLS. As will be shown, the algorithm itself is pretty simple. It boils down to regressing the transformed (and updated) outcome variable on the untransformed design matrix weighted by the variance of the transformed observations. This is done until we have convergence of the deviance function (twice the log-likelihood ratio of the current and previous estimates). The problem that I am running into with updating the binomial family to accept proportional data (ie., a vector of pairs (successes, total trials) instead of a vector of 1s and 0s for success or failure) is more mathematical than computational. I have either calculated the variance (and therefore the weights) incorrectly, or I am updating the outcome variable incorrectly. Of course, there's always the remote possibility that my data is not well behaved, but I don't think this is the case here.
More to come...
Generalized linear models was a topic that was completely foreign to me a few weeks ago, but after a little (okay a lot of) reading the approach seems almost natural. I have found the following references useful:
- Jeff Gill's Generalized Linear Models: A Unified Approach.
- James Hardin and Joseph Hilbe's Generalized Linear Models and Extensions, 2nd edition.
- P. McCullagh and John Nelder's Generalized Linear Models, 2nd edition.
The basic point of the generalized linear model is to extend the approach taken in classical linear regression to models that have more complex outcomes but ultimately share the linearity property. In this respect, GLM subsumes classical linear regression, probit and logit analysis, loglinear and multinomial response models, and some models that deal with survival data to name a few.
In my experience, I have found that econometrics is taught in a compartmentalized manner. This makes sense to a certain extent, as different estimators are tailored to particular problems and data. GLM on the other hand allows the use of a common a technique for obtaining parameter estimates so that it can be studied as a single technique rather than as a collection of distinct approaches.
If interested in my ramblings, you can find a draft of my notes as an introduction to GLM here, as Blogger does not support LaTeX... Please note that this is a preliminary and incomplete draft (corrections and clarifications are very welcome). One thing it could definitely use is some clarification by example. However, as I noted, I have run into a bit of a wall trying to extend the binomial family to accept a vector of proportional data, and this is my intended example to walk through the theory and algorithm, so... a subsequent post will have to lay this out once I've got it sorted myself.
Generally speaking, there are two basic algorithms for GLM estimation: one is a maximum likelihood optimization based on Newton's method the other is commonly refered to as iteratively (re)weighted least squares (IRLS or IWLS). Our implementation now only covers IRLS. As will be shown, the algorithm itself is pretty simple. It boils down to regressing the transformed (and updated) outcome variable on the untransformed design matrix weighted by the variance of the transformed observations. This is done until we have convergence of the deviance function (twice the log-likelihood ratio of the current and previous estimates). The problem that I am running into with updating the binomial family to accept proportional data (ie., a vector of pairs (successes, total trials) instead of a vector of 1s and 0s for success or failure) is more mathematical than computational. I have either calculated the variance (and therefore the weights) incorrectly, or I am updating the outcome variable incorrectly. Of course, there's always the remote possibility that my data is not well behaved, but I don't think this is the case here.
More to come...
Project Status
I have been making slow but steady progress on the NiPy models code. Right now for the midterm review, we have been focusing on design issues including the user interface and refactoring, test coverage/bug fixing, and some extensions for postestimation statistics. Other than this, I have spent the last month or so with anywhere from ten to fifteen stats, econometrics, or numerical linear algebra and optimization texts open on my desk.
The main estimators currently included in the code are generalized least squares, ordinary least squares, weighted least squares, autoregressive AR(p), generalized linear models (with several available distribution families and corresponding link functions), robust linear models, general additive models, and mixed effects models. The test coverage is starting to look pretty good, then there is just squashing the few remaining bugs and improving the postestimation statistics.
Some enhancements have also been made to the code. I have started to include some public domain or appropriately copyrighted datasets for testing purposes that could also be useful for examples and tutorials, so that every usage example doesn't have to start with generating your own random data. I have followed pretty closely to the datasets proposal in the Scikits Learn package.
We have also decided to break from the formula framework that is used in NiPy. It was in flux (being changed to take advantage of SymPy the last I heard) and is intended to be somewhat similar to the formula framework in R. In its place for now, I have written some convenience functions to append a constant to a design matrix or to handle categorical variables for estimation. For the moment, a typical model/estimator is used as
In [1]: from models.regression import OLS
In [2]: from models.datasets.longley.data import load
In [3]: from models.functions import add_constant
In [4]: data = load()
In [5]: data.exog = add_constant(data.exog)
In [6]: model = OLS(data.endog, data.exog)
In [7]: results = model.fit()
In [8]: results.params
Out[8]:
array([ 1.50618723e+01, -3.58191793e-02, -2.02022980e+00,
-1.03322687e+00, -5.11041057e-02, 1.82915146e+03,
-3.48225863e+06])
Barring any unforeseen difficulties, the models code should be available as a standalone package shortly after the midterm evaluation rapidly approaching in ten days. The second half of the summer will then be focused on optimizing the code, finalizing design issues, extending the models, and writing good documentation and tutorials so that the code can be included in SciPy!
The main estimators currently included in the code are generalized least squares, ordinary least squares, weighted least squares, autoregressive AR(p), generalized linear models (with several available distribution families and corresponding link functions), robust linear models, general additive models, and mixed effects models. The test coverage is starting to look pretty good, then there is just squashing the few remaining bugs and improving the postestimation statistics.
Some enhancements have also been made to the code. I have started to include some public domain or appropriately copyrighted datasets for testing purposes that could also be useful for examples and tutorials, so that every usage example doesn't have to start with generating your own random data. I have followed pretty closely to the datasets proposal in the Scikits Learn package.
We have also decided to break from the formula framework that is used in NiPy. It was in flux (being changed to take advantage of SymPy the last I heard) and is intended to be somewhat similar to the formula framework in R. In its place for now, I have written some convenience functions to append a constant to a design matrix or to handle categorical variables for estimation. For the moment, a typical model/estimator is used as
In [1]: from models.regression import OLS
In [2]: from models.datasets.longley.data import load
In [3]: from models.functions import add_constant
In [4]: data = load()
In [5]: data.exog = add_constant(data.exog)
In [6]: model = OLS(data.endog, data.exog)
In [7]: results = model.fit()
In [8]: results.params
Out[8]:
array([ 1.50618723e+01, -3.58191793e-02, -2.02022980e+00,
-1.03322687e+00, -5.11041057e-02, 1.82915146e+03,
-3.48225863e+06])
Barring any unforeseen difficulties, the models code should be available as a standalone package shortly after the midterm evaluation rapidly approaching in ten days. The second half of the summer will then be focused on optimizing the code, finalizing design issues, extending the models, and writing good documentation and tutorials so that the code can be included in SciPy!
Wednesday, July 1, 2009
Econometrics with Python
There is as yet no equivalent of R in applied econometrics. Therefore, the econometric community can still decide to go along the Python path.
That is Drs. Christine Choirat and Raffello Seri writing in the April issue of the Journal of Applied Econometrics. They have been kind enough to provide me with an ungated copy of their review, "Econometrics with Python." Mentioning the, quite frankly, redundant general programming functions and tools that had to be implemented for R, the authors make a nice case for Python as the programming language of choice for applied econometrics. The article provides a quick overview of some of the advantages of using Python and its many built-in libraries, extensions, and tools, gives some speed comparisons, and also mentions a few of the many tools out there in Python community for econometrics including RPy (RPy2 is now available), and of course NumPy and SciPy. Having spent the last week or more trying to master the basic syntax and usage of R, I very much sympathize with this position. The one complaint I hear most often from my fellow students is that Python is not an industry standard. I hope this can change and is changing, because it's much more of a pleasure to work with Python than the alternatives and that makes for increased productivity.
Monday, June 15, 2009
Legendre on Least Squares
I found the epigraph to Åke Björk's Numerical Methods for Least Squares Problems to be a nice intersection of my interests.
Adrien Marie Legendre, Nouvelles méthodes pour la détermination des orbites des comètes. Appendice. Paris, 1805.
De tous les principes qu'on peut proposer pour cet object, je pense qu'il n'en est pas de plus general, de plus exact, ni d'une application plus facile que celui qui consiste à rendre minimum la somme de carrés des erreurs.
Of all the principles that can be proposed, I think there is none more general, more exact, or of an easier application than that which consists of rendering the sum of squared errors a minimum.
Adrien Marie Legendre, Nouvelles méthodes pour la détermination des orbites des comètes. Appendice. Paris, 1805.
Friday, June 12, 2009
Design Issues: Understanding Python's super
The current statistical models package is housed in the NiPy, Neuroimaging in Python, project. Right now, it is designed to rely on Python's built-in super to handle class inheritance. This post will dig a little more into the super function and what it means for the design of the project and future extensions. Note that there are plenty of good places to learn about super and that this post is to help me as much as anyone else. [*Edit: With this in mind, I direct you to Things to Know about Python Super if you really want a deeper and correct understanding of super. This post is mainly a heuristic approach that has helped me in understanding basic usage of super.] You can find the documentation for super here. If this is a bit confusing, it will, I hope, become clearer after I demonstrate the usage.
First, let's take a look at how super actually works for the simple case of single inheritance (right now, we are not planning on using multiple inheritance in the project) and an __init__ chain (note that super can call any of its parent class's methods, but using __init__ is my current use case).
The following examples were adapted from some code provided by mentors (thank you!).
class A(object):
def __init__(self, a):
self.a = a
print 'executing A().__init__'
class B(A):
def __init__(self, a):
self.ab = a*2
print 'executing B().__init__'
super(B,self).__init__(a)
class C(B):
def __init__(self, a):
self.ac = a*3
print 'executing C().__init__'
super(C,self).__init__(a)
Now let's have a look at creating an instance of C.
In [2]: cinst = C(10)
executing C().__init__
executing B().__init__
executing A().__init__
In [3]: vars(cinst)
Out[3]: {'a': 10, 'ab': 20, 'ac': 30}
That seems simple enough. Creating an instance of C with a = 10 will also give C the attributes of B(10) and A(10). This means our one instance of C has three attributes: cinst.ac, cinst.ab, cinst.a. The latter two were created by its parent classes (or superclasses) __init__ method. Note that A is also a new-style class. It subclasses the 'object' type.
The actual calls to super pass the generic class 'C' and a handle to that class 'self', which is 'cinst' in our case. Super returns the literal parent of the class instance C since we passed it 'self'. It should be noted that A and B were created when we initialized cinst and are, therefore, 'bound' class objects (bound to cinst in memory through the actual instance of class C) and not referring to the class A and class B instructions defined at the interpreter (assuming you are typing along at the interpreter).
Okay now let's define a few more classes to look briefly at multiple inheritance.
class D(A):
def __init__(self, a):
self.ad = a*4
print 'executing D().__init__'
# if super is commented out then __init__ chain ends
#super(D,self).__init__(a)
class E(D):
def __init__(self, a):
self.ae = a*5
print 'executing E().__init__'
super(E,self).__init__(a)
Note that the call to super in D is commented out. This breaks the __init__ chain.
In [4]: einst = E(10)
executing E().__init__
executing D().__init__
In [5]: vars(einst)
Out[5]: {'ad': 40, 'ae': 50}
If we uncomment the super in D, we get as we would expect
In [6]: einst = E(10)
executing E().__init__
executing D().__init__
executing A().__init__
In [7]: vars(einst)
Out[7]: {'a': 10, 'ad': 40, 'ae': 50}
Ok that's pretty straightforward. In this way super is used to pass off something to its parent class. For instance, say we have a little more realistic example and the instance of C takes some timeseries data that exhibits serial correlation. Then we can have C correct for the covariance structure of the data and "pass it up" to B where B can then perform OLS on our data now that it meets the assumptions of OLS. Further B can pass this data to A and return some descriptive statistics for our data. But remember these are 'bound' class objects, so they're all attributes to our original instance of C. Neat huh? Okay, now let's look at a pretty simple example of multiple inheritance.
class F(C,E):
def __init__(self, a):
self.af = a*6
print 'executing F().__init__'
super(F,self).__init__(a)
For this example we are using the class of D, that has super commented out.
In [8]: finst = F(10)
executing F().__init__
executing C().__init__
executing B().__init__
executing E().__init__
executing D().__init__
In [8]: vars(finst)
Out[8]: {'ab': 20, 'ac': 30, 'ad': 40, 'ae': 50, 'af': 60}
The first time I saw this gave me pause. Why isn't there an finst.a? I was expecting the MRO to go F -> C -> B -> A -> E -> D -> A. Let's take a closer look. The F class has multiple inheritance. It inherits from both C and E. We can see F's method resolution order by doing
In [9]: F.__mro__
Out[9]:
(<class '__main__.F'>,
<class '__main__.C'>,
<class '__main__.B'>,
<class '__main__.E'>,
<class '__main__.D'>,
<class '__main__.A'>,
<type 'object'>)
Okay, so we can see that for F A is a subclass of D but not B. But why?
In [10]: A.__subclasses__()
Out[10]: [<class '__main__.B'>, <class '__main__.D'>]
The reason is that A does not have a call to super, so the chain doesn't exist here. When you instantiate F, the hierarchy goes F -> C -> B -> E -> D -> A. The reason that it goes from B -> E is because A does not have a call to super, so it can't pass anything to E (It couldn't pass anything to E because the object.__init__ doesn't take a parameter "a" and because you cannot have a MRO F -> C -> B -> A -> E -> D -> A as this is inconsistent and will give an error!), so A does not cause a problem and the chain ends after D (remember that D's super is commented out, but if it were not then there would be finst.a = 10 as expected). Whew.
I'm sure you're thinking "Oh that's (relatively) easy. I'm ready to go crazy with super." But there are a number of things must keep in mind when using super, which makes it necessary for the users of super to proceed carefully.
1. super() only works with new-style classes. You can read more about classic/old-style vs new-style classes here. From there you can click through or just go here for more information on new-style classes. Therefore, you must know that the base classes are new-style. This isn't a problem for our project right now, because I have access to all of the base classes.
2. Subclasses must use super if their superclasses do. This is why the user of super must be well-documented. If we have to classes A and B that both use super and a class C that inherits from them, but does not know about super then we will have a problem. Consider the slightly different case
class A(object):
def __init__(self):
print "executing A().__init__"
super(A, self).__init__()
class B(object):
def __init__(self):
print "executing B().__init__"
super(B, self).__init__()
class C(A,B):
def __init__(self):
print "executing C().__init__"
A.__init__(self)
B.__init__(self)
# super(C, self).__init__()
Say class C was defined by someone who couldn't see class A and B, then they wouldn't know about super. Now if we do
In [11]: C.__mro__
Out[11]:
(<class '__main__.C'>,
<class '__main__.A'>,
<class '__main__.B'>,
<type 'object'>)
In [12]: c = C()
executing C().__init__
executing A().__init__
executing B().__init__
executing B().__init__
B got called twice, but by now this should be expected. A's super calls __init__ on the next object in the MRO which is B (it works this time unlike above because there is no parameter passed with __init__), then C explicitly calls B again.
If we uncomment super and comment out the calls to the parent __init__ methods in C then this works as expected.
3. Superclasses probably should use super if their subclasses do.
We saw this earlier with class D's super call commented out. Note also that A does not have a call to super. The last class in the MRO does not need super *if* there is only one such class at the end.
4. Classes must have the exact same call signature.
This should be obvious but is important for people to be able to subclass. It is possible however for subclasses to add additional arguments so *args and **kwargs should be probably always be included in the methods that are accessible to subclasses.
5. Because of these last three points, the use of super must be explicitly documented, as it has become a part of the interface to our classes.
First, let's take a look at how super actually works for the simple case of single inheritance (right now, we are not planning on using multiple inheritance in the project) and an __init__ chain (note that super can call any of its parent class's methods, but using __init__ is my current use case).
The following examples were adapted from some code provided by mentors (thank you!).
class A(object):
def __init__(self, a):
self.a = a
print 'executing A().__init__'
class B(A):
def __init__(self, a):
self.ab = a*2
print 'executing B().__init__'
super(B,self).__init__(a)
class C(B):
def __init__(self, a):
self.ac = a*3
print 'executing C().__init__'
super(C,self).__init__(a)
Now let's have a look at creating an instance of C.
In [2]: cinst = C(10)
executing C().__init__
executing B().__init__
executing A().__init__
In [3]: vars(cinst)
Out[3]: {'a': 10, 'ab': 20, 'ac': 30}
That seems simple enough. Creating an instance of C with a = 10 will also give C the attributes of B(10) and A(10). This means our one instance of C has three attributes: cinst.ac, cinst.ab, cinst.a. The latter two were created by its parent classes (or superclasses) __init__ method. Note that A is also a new-style class. It subclasses the 'object' type.
The actual calls to super pass the generic class 'C' and a handle to that class 'self', which is 'cinst' in our case. Super returns the literal parent of the class instance C since we passed it 'self'. It should be noted that A and B were created when we initialized cinst and are, therefore, 'bound' class objects (bound to cinst in memory through the actual instance of class C) and not referring to the class A and class B instructions defined at the interpreter (assuming you are typing along at the interpreter).
Okay now let's define a few more classes to look briefly at multiple inheritance.
class D(A):
def __init__(self, a):
self.ad = a*4
print 'executing D().__init__'
# if super is commented out then __init__ chain ends
#super(D,self).__init__(a)
class E(D):
def __init__(self, a):
self.ae = a*5
print 'executing E().__init__'
super(E,self).__init__(a)
Note that the call to super in D is commented out. This breaks the __init__ chain.
In [4]: einst = E(10)
executing E().__init__
executing D().__init__
In [5]: vars(einst)
Out[5]: {'ad': 40, 'ae': 50}
If we uncomment the super in D, we get as we would expect
In [6]: einst = E(10)
executing E().__init__
executing D().__init__
executing A().__init__
In [7]: vars(einst)
Out[7]: {'a': 10, 'ad': 40, 'ae': 50}
Ok that's pretty straightforward. In this way super is used to pass off something to its parent class. For instance, say we have a little more realistic example and the instance of C takes some timeseries data that exhibits serial correlation. Then we can have C correct for the covariance structure of the data and "pass it up" to B where B can then perform OLS on our data now that it meets the assumptions of OLS. Further B can pass this data to A and return some descriptive statistics for our data. But remember these are 'bound' class objects, so they're all attributes to our original instance of C. Neat huh? Okay, now let's look at a pretty simple example of multiple inheritance.
class F(C,E):
def __init__(self, a):
self.af = a*6
print 'executing F().__init__'
super(F,self).__init__(a)
For this example we are using the class of D, that has super commented out.
In [8]: finst = F(10)
executing F().__init__
executing C().__init__
executing B().__init__
executing E().__init__
executing D().__init__
In [8]: vars(finst)
Out[8]: {'ab': 20, 'ac': 30, 'ad': 40, 'ae': 50, 'af': 60}
The first time I saw this gave me pause. Why isn't there an finst.a? I was expecting the MRO to go F -> C -> B -> A -> E -> D -> A. Let's take a closer look. The F class has multiple inheritance. It inherits from both C and E. We can see F's method resolution order by doing
In [9]: F.__mro__
Out[9]:
(<class '__main__.F'>,
<class '__main__.C'>,
<class '__main__.B'>,
<class '__main__.E'>,
<class '__main__.D'>,
<class '__main__.A'>,
<type 'object'>)
Okay, so we can see that for F A is a subclass of D but not B. But why?
In [10]: A.__subclasses__()
Out[10]: [<class '__main__.B'>, <class '__main__.D'>]
The reason is that A does not have a call to super, so the chain doesn't exist here. When you instantiate F, the hierarchy goes F -> C -> B -> E -> D -> A. The reason that it goes from B -> E is because A does not have a call to super, so it can't pass anything to E (It couldn't pass anything to E because the object.__init__ doesn't take a parameter "a" and because you cannot have a MRO F -> C -> B -> A -> E -> D -> A as this is inconsistent and will give an error!), so A does not cause a problem and the chain ends after D (remember that D's super is commented out, but if it were not then there would be finst.a = 10 as expected). Whew.
I'm sure you're thinking "Oh that's (relatively) easy. I'm ready to go crazy with super." But there are a number of things must keep in mind when using super, which makes it necessary for the users of super to proceed carefully.
1. super() only works with new-style classes. You can read more about classic/old-style vs new-style classes here. From there you can click through or just go here for more information on new-style classes. Therefore, you must know that the base classes are new-style. This isn't a problem for our project right now, because I have access to all of the base classes.
2. Subclasses must use super if their superclasses do. This is why the user of super must be well-documented. If we have to classes A and B that both use super and a class C that inherits from them, but does not know about super then we will have a problem. Consider the slightly different case
class A(object):
def __init__(self):
print "executing A().__init__"
super(A, self).__init__()
class B(object):
def __init__(self):
print "executing B().__init__"
super(B, self).__init__()
class C(A,B):
def __init__(self):
print "executing C().__init__"
A.__init__(self)
B.__init__(self)
# super(C, self).__init__()
Say class C was defined by someone who couldn't see class A and B, then they wouldn't know about super. Now if we do
In [11]: C.__mro__
Out[11]:
(<class '__main__.C'>,
<class '__main__.A'>,
<class '__main__.B'>,
<type 'object'>)
In [12]: c = C()
executing C().__init__
executing A().__init__
executing B().__init__
executing B().__init__
B got called twice, but by now this should be expected. A's super calls __init__ on the next object in the MRO which is B (it works this time unlike above because there is no parameter passed with __init__), then C explicitly calls B again.
If we uncomment super and comment out the calls to the parent __init__ methods in C then this works as expected.
3. Superclasses probably should use super if their subclasses do.
We saw this earlier with class D's super call commented out. Note also that A does not have a call to super. The last class in the MRO does not need super *if* there is only one such class at the end.
4. Classes must have the exact same call signature.
This should be obvious but is important for people to be able to subclass. It is possible however for subclasses to add additional arguments so *args and **kwargs should be probably always be included in the methods that are accessible to subclasses.
5. Because of these last three points, the use of super must be explicitly documented, as it has become a part of the interface to our classes.
Saturday, May 9, 2009
Working with Data: Record Arrays
A few posts are going to be directed at people who are not as familiar with Python and/or SciPy. I will try to assume as little as possible about what the user knows. Over the summer these types of posts might be put together as a larger tutorial.
One of the goals of the SciPy stats project is to provide a transparent way to work with the different arrays types. Towards this end, we are going to work with some data as record arrays (note: right now this is only supported for files containing ASCII characters. If this is not the case, you must do some data cleaning beforehand with Python).
We have four data files: a comma-delimited .csv file with variable labels (headers) with all numeric data (here, info here), a comma-delimited file with headers with a mix of numeric and string variables (here, info here), this same file without headers (here), and this same file without headers that is tab delimited (here).
First, we are going to put the data into record arrays. Record arrays are simply structured arrays that allow access to the data through attribute access. If you are interested, you can have a look here for a bit more about the basic data types in NumPy. Then see here for more on structured and record arrays (and why access to record arrays is slower) and here for more details on creating and working with record arrays.
First download the data files and put them in a directory. I am using Linux, so my directory paths will look a little different than those using Windows, but all of the other details should be the same.
>>>import numpy as np
>>>educ_dta=np.recfromcsv('/home/skipper/scipystats/scipystats/data/educ_data.csv')
We now have our data in a record array. We can take a closer look at what's going on by typing
>>>educ_dta.dtype
dtype([('ahe', '<f8'), ('female', '<i8'), ('ne', '<i8'), ('midwest', '<i8'), ('south', '<i8'), ('west', '<i8'), ('race', '<i8'), ('yrseduc', '<i8'), ('ba', '<i8'), ('hsdipl', '<i8'), ('age', '<i8')])
>>>educ_dta.dtype.names
('ahe', 'female', 'ne', 'midwest', 'south', 'west', 'race', 'yrseduc', 'ba', 'hsdipl', 'age')
>>>educ_dta.ahe
array([ 12.5 , 6.25, 17.31, ..., 9.13, 11.11, 14.9 ])
>>>educ_dta['ahe']
array([ 12.5 , 6.25, 17.31, ..., 9.13, 11.11, 14.9 ])
We can do the same for each of our other data files. First we have another dataset that contains a string variable for the state name. We proceed just as above, though the string variables will have to be handled differently when used in a statistical model. Then we have the same file, but without any headers information. Last we have a file without headers that is tab-delimited.
>>>gun_dta=np.recfromcsv('/home/skipper/scipystats/scipystats/data/handguns_data.csv')
>>>gun_dta_nh=np.recfromcsv('/home/skipper/scipystats/scipystats/data/handguns_data_noheaders.csv', names=None)
>>>gun_dta_tnh=np.recfromtxt('home/skipper/scipystats/scipystats/data/handguns_data_tab_noheaders', names=None, delimiter='\t')
You can learn more about the possibilities for loading data into arrays here, or by having a look at the doc string for np.genfromtxt in the Python interpreter. All of the functions loadtxt, recfromcsv, recfromtxt, etc. use genfromtxt they just have different default values for the keyword arguments.
One of the goals of the SciPy stats project is to provide a transparent way to work with the different arrays types. Towards this end, we are going to work with some data as record arrays (note: right now this is only supported for files containing ASCII characters. If this is not the case, you must do some data cleaning beforehand with Python).
We have four data files: a comma-delimited .csv file with variable labels (headers) with all numeric data (here, info here), a comma-delimited file with headers with a mix of numeric and string variables (here, info here), this same file without headers (here), and this same file without headers that is tab delimited (here).
First, we are going to put the data into record arrays. Record arrays are simply structured arrays that allow access to the data through attribute access. If you are interested, you can have a look here for a bit more about the basic data types in NumPy. Then see here for more on structured and record arrays (and why access to record arrays is slower) and here for more details on creating and working with record arrays.
First download the data files and put them in a directory. I am using Linux, so my directory paths will look a little different than those using Windows, but all of the other details should be the same.
>>>import numpy as np
>>>educ_dta=np.recfromcsv('/home/skipper/scipystats/scipystats/data/educ_data.csv')
We now have our data in a record array. We can take a closer look at what's going on by typing
>>>educ_dta.dtype
dtype([('ahe', '<f8'), ('female', '<i8'), ('ne', '<i8'), ('midwest', '<i8'), ('south', '<i8'), ('west', '<i8'), ('race', '<i8'), ('yrseduc', '<i8'), ('ba', '<i8'), ('hsdipl', '<i8'), ('age', '<i8')])
>>>educ_dta.dtype.names
('ahe', 'female', 'ne', 'midwest', 'south', 'west', 'race', 'yrseduc', 'ba', 'hsdipl', 'age')
>>>educ_dta.ahe
array([ 12.5 , 6.25, 17.31, ..., 9.13, 11.11, 14.9 ])
>>>educ_dta['ahe']
array([ 12.5 , 6.25, 17.31, ..., 9.13, 11.11, 14.9 ])
We can do the same for each of our other data files. First we have another dataset that contains a string variable for the state name. We proceed just as above, though the string variables will have to be handled differently when used in a statistical model. Then we have the same file, but without any headers information. Last we have a file without headers that is tab-delimited.
>>>gun_dta=np.recfromcsv('/home/skipper/scipystats/scipystats/data/handguns_data.csv')
>>>gun_dta_nh=np.recfromcsv('/home/skipper/scipystats/scipystats/data/handguns_data_noheaders.csv', names=None)
>>>gun_dta_tnh=np.recfromtxt('home/skipper/scipystats/scipystats/data/handguns_data_tab_noheaders', names=None, delimiter='\t')
You can learn more about the possibilities for loading data into arrays here, or by having a look at the doc string for np.genfromtxt in the Python interpreter. All of the functions loadtxt, recfromcsv, recfromtxt, etc. use genfromtxt they just have different default values for the keyword arguments.
Friday, April 24, 2009
Getting Started with GSoC and SciPy
I'm trying to resist making the obligatory "hello world" post here. The best I can do is only mentioning the urge.
First, a little bit about myself. My name is Skipper Seabold. I am finishing my first year as a PhD student in economics at American University in Washington, DC, and I have recently been accepted to the Google Summer of Code 2009 to work on the SciPy project. I have been a computer hardware and programming hobbyist since my middle school days. I have built my computers my whole life and back in high school tinkered around with Visual Basic (Apps for AOL 3.0 on Windows 3.x and Windows 95 anyone?), Turbo Pascal, C++, and Java mostly in the context of coursework. Two years ago I was introduced to the Python programming language, and I haven't looked back. Needless to say I'm very happy to have two of my interests, economics and programming, overlap.
This is where SciPy comes in. For those who are unfamiliar with SciPy, I direct you to the homepage here. In short, SciPy is an open source library of algorithms for numerical analysis for those working in engineering or the sciences more broadly defined. The SciPy library depends on NumPy. The Tentative NumPy Tutorial is a good place to start learning about the capabilities of NumPy. And likewise, the Getting Started page has plenty of resources to introduce you to the power of SciPy. In particular the tutorials, documentation, and cookbook are good to look at.
What I will be working on this summer is providing a consistent user interface for statistical models and appropriate statistical tests in SciPy similar to those found in other statistics/econometric software packages. I will also provide a unified development framework for those who would like to add to this effort in the future. Updates may be less regular over the next few weeks, but check here for at least weekly updates on the work over the summer.
First, a little bit about myself. My name is Skipper Seabold. I am finishing my first year as a PhD student in economics at American University in Washington, DC, and I have recently been accepted to the Google Summer of Code 2009 to work on the SciPy project. I have been a computer hardware and programming hobbyist since my middle school days. I have built my computers my whole life and back in high school tinkered around with Visual Basic (Apps for AOL 3.0 on Windows 3.x and Windows 95 anyone?), Turbo Pascal, C++, and Java mostly in the context of coursework. Two years ago I was introduced to the Python programming language, and I haven't looked back. Needless to say I'm very happy to have two of my interests, economics and programming, overlap.
This is where SciPy comes in. For those who are unfamiliar with SciPy, I direct you to the homepage here. In short, SciPy is an open source library of algorithms for numerical analysis for those working in engineering or the sciences more broadly defined. The SciPy library depends on NumPy. The Tentative NumPy Tutorial is a good place to start learning about the capabilities of NumPy. And likewise, the Getting Started page has plenty of resources to introduce you to the power of SciPy. In particular the tutorials, documentation, and cookbook are good to look at.
What I will be working on this summer is providing a consistent user interface for statistical models and appropriate statistical tests in SciPy similar to those found in other statistics/econometric software packages. I will also provide a unified development framework for those who would like to add to this effort in the future. Updates may be less regular over the next few weeks, but check here for at least weekly updates on the work over the summer.
Subscribe to:
Comments (Atom)
